One-to-n alignments
Command: compare-matrices -v 1 -mode matches -format1 transfac -file1 $RSAT/public_html/tmp/wwwrun/2013/01/11/peak-motifs.2013-01-11.110705_2013-01-11.110705_PdhmNx/results/discovered_motifs/positions_7nt_m3/peak-motifs_positions_7nt_m3.tf -format2 tf -file2 $RSAT/public_html/data/motif_databases/JASPAR/jaspar_core_vertebrates_2009_10.tf -mode matches -DR -uth offset_rank 1 -lth w 5 -lth Wr 0.3 -lth cor 0.75 -lth Ncor 0.4 -return matrix_name,matrix_id,cor,Ncor,logoDP,NIcor,NsEucl,SSD,NSW,match_rank,width,strand,offset,consensus,alignments_1ton -sort Ncor -o $RSAT/public_html/tmp/wwwrun/2013/01/11/peak-motifs.2013-01-11.110705_2013-01-11.110705_PdhmNx/results/discovered_motifs/positions_7nt_m3/peak-motifs_positions_7nt_m3_vs_db_jaspar_core_vertebrates
One-to-n matrix alignment; reference matrix: positions_7nt_m3_shift0 ; 6 matrices ; sort_field=rank_mean
| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SSD |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSSD |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
| positions_7nt_m3_shift0 (positions_7nt_m3) |
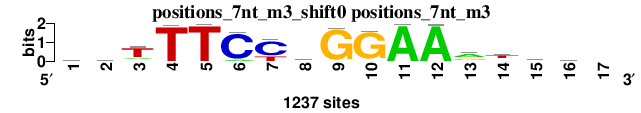 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; positions_7nt_m3; m=0 (reference); ncol1=16; shift=0; ncol=17; srTTTCYwGGAArtsr-
; Alignment reference
a 225 433 191 3 3 105 1 380 30 39 1231 1228 634 142 240 404 0
c 372 246 86 4 3 1125 892 153 7 2 2 3 44 216 326 279 0
g 375 329 66 7 2 3 1 306 1196 1157 1 2 411 156 453 317 0
t 265 229 894 1223 1229 4 343 398 4 39 3 4 148 723 218 237 0
|
| MA0137.2_rc_shift0 (STAT1_rc) |
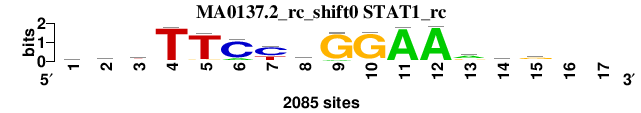 |
0.975 |
0.914 |
8.663 |
0.878 |
0.974 |
0.307 |
0.990 |
1 |
1 |
2 |
1 |
1 |
1 |
1 |
1.143 |
1 |
; positions_7nt_m3 versus MA0137.2_rc (STAT1_rc); m=1/5; ncol2=15; w=15; offset=0; strand=R; shift=0; score= 1.1429; gryTTCydGGAArtg--
; cor=0.975; Ncor=0.914; logoDP=8.663; NIcor=0.878; NsEucl=0.974; SSD=0.307; NSW=0.990; rcor=1; rNcor=1; rlogoDP=2; rNIcor=1; rNsEucl=1; rSSD=1; rNSW=1; rank_mean=1.143; match_rank=1
a 431 714 315 15 32 302 66 680 155 44 2023 2038 1112 446 378 0 0
c 425 270 552 23 29 1700 1292 115 7 14 38 11 144 279 415 0 0
g 804 760 263 17 124 30 31 762 1900 1921 14 22 574 496 1076 0 0
t 417 336 954 2030 1900 53 696 528 23 106 8 10 251 859 208 0 0
|
| MA0144.1_shift3 (Stat3) |
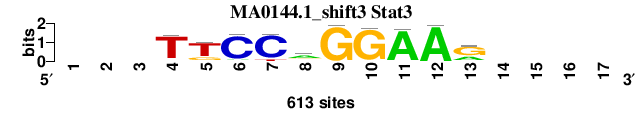 |
0.963 |
0.602 |
9.169 |
0.595 |
0.956 |
0.390 |
0.980 |
2 |
2 |
1 |
2 |
2 |
2 |
2 |
1.857 |
2 |
; positions_7nt_m3 versus MA0144.1 (Stat3); m=2/5; ncol2=10; w=10; offset=3; strand=D; shift=3; score= 1.8571; ---TTCCaGGAAr----
; cor=0.963; Ncor=0.602; logoDP=9.169; NIcor=0.595; NsEucl=0.956; SSD=0.390; NSW=0.980; rcor=2; rNcor=2; rlogoDP=1; rNIcor=2; rNsEucl=2; rSSD=2; rNSW=2; rank_mean=1.857; match_rank=2
a 0 0 0 20 13 38 6 321 8 6 585 606 191 0 0 0 0
c 0 0 0 19 10 552 541 21 0 2 21 1 15 0 0 0 0
g 0 0 0 25 129 9 1 148 605 592 7 5 393 0 0 0 0
t 0 0 0 549 461 14 65 123 0 13 0 1 14 0 0 0 0
|
| MA0156.1_shift6 (FEV) |
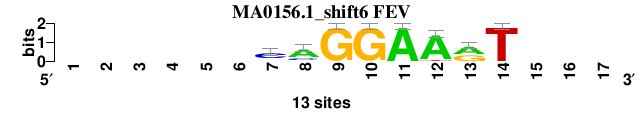 |
0.925 |
0.462 |
6.770 |
0.460 |
0.929 |
0.646 |
0.960 |
3 |
5 |
4 |
4 |
3 |
3 |
3 |
3.571 |
3 |
; positions_7nt_m3 versus MA0156.1 (FEV); m=3/5; ncol2=8; w=8; offset=6; strand=D; shift=6; score= 3.5714; ------CAGGAArT---
; cor=0.925; Ncor=0.462; logoDP=6.770; NIcor=0.460; NsEucl=0.929; SSD=0.646; NSW=0.960; rcor=3; rNcor=5; rlogoDP=4; rNIcor=4; rNsEucl=3; rSSD=3; rNSW=3; rank_mean=3.571; match_rank=3
a 0 0 0 0 0 0 2 9 0 0 13 12 7 0 0 0 0
c 0 0 0 0 0 0 9 3 0 0 0 0 0 0 0 0 0
g 0 0 0 0 0 0 1 1 13 13 0 0 6 0 0 0 0
t 0 0 0 0 0 0 1 0 0 0 0 1 0 13 0 0 0
|
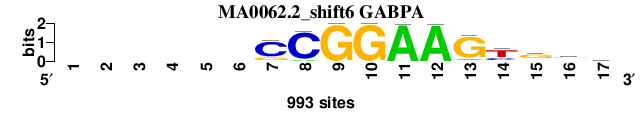
| MA0062.2_shift6 (GABPA) |
 |
0.822 |
0.483 |
7.163 |
0.488 |
0.910 |
1.621 |
0.919 |
5 |
3 |
3 |
3 |
5 |
5 |
5 |
4.143 |
4 |
; positions_7nt_m3 versus MA0062.2 (GABPA); m=4/5; ncol2=11; w=10; offset=6; strand=D; shift=6; score= 4.1429; ------CCGGAAGygvc
; cor=0.822; Ncor=0.483; logoDP=7.163; NIcor=0.488; NsEucl=0.910; SSD=1.621; NSW=0.919; rcor=5; rNcor=3; rlogoDP=3; rNIcor=3; rNsEucl=5; rSSD=5; rNSW=5; rank_mean=4.143; match_rank=4
a 0 0 0 0 0 0 32 70 0 0 991 989 94 56 154 264 233
c 0 0 0 0 0 0 768 914 0 0 1 2 32 261 137 262 357
g 0 0 0 0 0 0 188 4 990 991 1 0 866 37 603 415 224
t 0 0 0 0 0 0 1 1 1 0 0 2 0 637 96 49 176
|
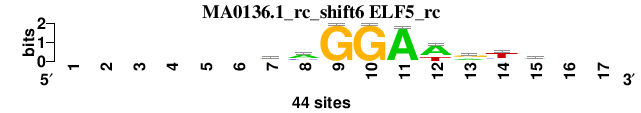
| MA0136.1_rc_shift6 (ELF5_rc) |
 |
0.834 |
0.469 |
0.177 |
-0.102 |
0.917 |
1.111 |
0.938 |
4 |
4 |
5 |
5 |
4 |
4 |
4 |
4.286 |
5 |
; positions_7nt_m3 versus MA0136.1_rc (ELF5_rc); m=5/5; ncol2=9; w=9; offset=6; strand=R; shift=6; score= 4.2857; ------mmGGAwrtw--
; cor=0.834; Ncor=0.469; logoDP=0.177; NIcor=-0.102; NsEucl=0.917; SSD=1.111; NSW=0.938; rcor=4; rNcor=4; rlogoDP=5; rNIcor=5; rNsEucl=4; rSSD=4; rNSW=4; rank_mean=4.286; match_rank=5
a 0 0 0 0 0 0 21 26 0 0 43 29 17 5 20 0 0
c 0 0 0 0 0 0 11 11 0 0 0 1 5 7 3 0 0
g 0 0 0 0 0 0 8 3 44 44 1 0 20 3 9 0 0
t 0 0 0 0 0 0 4 4 0 0 0 14 2 29 12 0 0
|





