| Matrix name | Aligned logos | cor |
Ncor |
logoDP |
NIcor |
NsEucl |
SSD |
NSW |
rcor |
rNcor |
rlogoDP |
rNIcor |
rNsEucl |
rSSD |
rNSW |
rank_mean |
match_rank |
Aligned matrices |
|---|
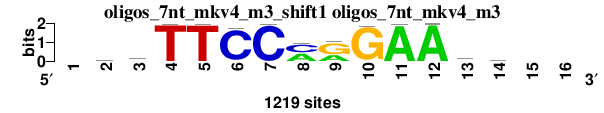
| oligos_7nt_mkv4_m3_shift1 (oligos_7nt_mkv4_m3) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
; oligos_7nt_mkv4_m3; m=0 (reference); ncol1=13; shift=1; ncol=16; -ryTTCCmrGAAry--
; Alignment reference
a 0 381 217 0 4 8 1 597 450 1 1213 1217 492 233 0 0
c 0 271 347 0 0 1168 1214 612 1 0 3 2 185 354 0 0
g 0 337 158 3 1 0 4 10 768 1195 0 0 333 244 0 0
t 0 230 497 1216 1214 43 0 0 0 23 3 0 209 388 0 0
|
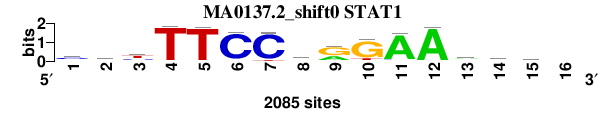
| MA0137.2_shift0 (STAT1) |
 |
0.972 |
0.842 |
9.634 |
0.851 |
0.969 |
0.329 |
0.987 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1.000 |
1 |
; oligos_7nt_mkv4_m3 versus MA0137.2 (STAT1); m=1/8; ncol2=15; w=13; offset=-1; strand=D; shift=0; score= 1; cayTTCChrGAAryc-
; cor=0.972; Ncor=0.842; logoDP=9.634; NIcor=0.851; NsEucl=0.969; SSD=0.329; NSW=0.987; rcor=1; rNcor=1; rlogoDP=1; rNIcor=1; rNsEucl=1; rSSD=1; rNSW=1; rank_mean=1.000; match_rank=1
a 208 859 251 10 8 106 23 528 696 53 1900 2030 954 336 417 0
c 1076 496 574 22 14 1921 1900 762 31 30 124 17 263 760 804 0
g 415 279 144 11 38 14 7 115 1292 1700 29 23 552 270 425 0
t 378 446 1112 2038 2023 44 155 680 66 302 32 15 315 714 431 0
|
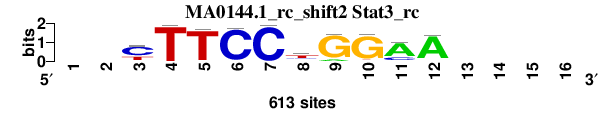
| MA0144.1_rc_shift2 (Stat3_rc) |
 |
0.922 |
0.709 |
4.339 |
0.204 |
0.934 |
0.861 |
0.957 |
2 |
2 |
6 |
6 |
2 |
3 |
2 |
3.286 |
2 |
; oligos_7nt_mkv4_m3 versus MA0144.1_rc (Stat3_rc); m=2/8; ncol2=10; w=10; offset=1; strand=R; shift=2; score= 3.2857; --yTTCCtGGAA----
; cor=0.922; Ncor=0.709; logoDP=4.339; NIcor=0.204; NsEucl=0.934; SSD=0.861; NSW=0.957; rcor=2; rNcor=2; rlogoDP=6; rNIcor=6; rNsEucl=2; rSSD=3; rNSW=2; rank_mean=3.286; match_rank=2
a 0 0 14 1 0 13 0 123 65 14 461 549 0 0 0 0
c 0 0 393 5 7 592 605 148 1 9 129 25 0 0 0 0
g 0 0 15 1 21 2 0 21 541 552 10 19 0 0 0 0
t 0 0 191 606 585 6 8 321 6 38 13 20 0 0 0 0
|
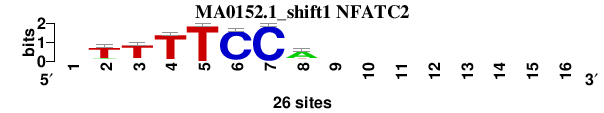
| MA0152.1_shift1 (NFATC2) |
 |
0.875 |
0.471 |
6.581 |
0.469 |
0.904 |
0.901 |
0.936 |
4 |
4 |
3 |
3 |
3 |
4 |
4 |
3.571 |
3 |
; oligos_7nt_mkv4_m3 versus MA0152.1 (NFATC2); m=3/8; ncol2=7; w=7; offset=0; strand=D; shift=1; score= 3.5714; -TTTTCCA--------
; cor=0.875; Ncor=0.471; logoDP=6.581; NIcor=0.469; NsEucl=0.904; SSD=0.901; NSW=0.936; rcor=4; rNcor=4; rlogoDP=3; rNIcor=3; rNsEucl=3; rSSD=4; rNSW=4; rank_mean=3.571; match_rank=3
a 0 3 1 1 0 1 0 18 0 0 0 0 0 0 0 0
c 0 1 2 1 0 25 26 3 0 0 0 0 0 0 0 0
g 0 2 2 0 0 0 0 1 0 0 0 0 0 0 0 0
t 0 20 21 24 26 0 0 4 0 0 0 0 0 0 0 0
|
| MA0156.1_shift6 (FEV) |
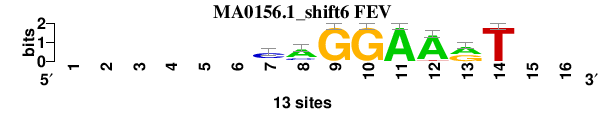 |
0.846 |
0.521 |
6.282 |
0.484 |
0.901 |
1.264 |
0.921 |
5 |
3 |
4 |
2 |
5 |
6 |
6 |
4.429 |
4 |
; oligos_7nt_mkv4_m3 versus MA0156.1 (FEV); m=4/8; ncol2=8; w=8; offset=5; strand=D; shift=6; score= 4.4286; ------CAGGAArT--
; cor=0.846; Ncor=0.521; logoDP=6.282; NIcor=0.484; NsEucl=0.901; SSD=1.264; NSW=0.921; rcor=5; rNcor=3; rlogoDP=4; rNIcor=2; rNsEucl=5; rSSD=6; rNSW=6; rank_mean=4.429; match_rank=4
a 0 0 0 0 0 0 2 9 0 0 13 12 7 0 0 0
c 0 0 0 0 0 0 9 3 0 0 0 0 0 0 0 0
g 0 0 0 0 0 0 1 1 13 13 0 0 6 0 0 0
t 0 0 0 0 0 0 1 0 0 0 0 1 0 13 0 0
|
| MA0098.1_rc_shift7 (ETS1_rc) |
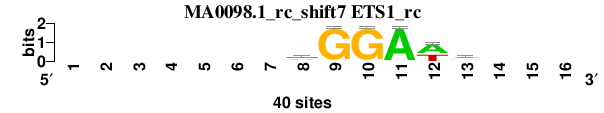 |
0.876 |
0.404 |
0.152 |
-0.109 |
0.904 |
0.662 |
0.945 |
3 |
8 |
8 |
7 |
4 |
2 |
3 |
5.000 |
5 |
; oligos_7nt_mkv4_m3 versus MA0098.1_rc (ETS1_rc); m=5/8; ncol2=6; w=6; offset=6; strand=R; shift=7; score= 5; -------mGGAwr---
; cor=0.876; Ncor=0.404; logoDP=0.152; NIcor=-0.109; NsEucl=0.904; SSD=0.662; NSW=0.945; rcor=3; rNcor=8; rlogoDP=8; rNIcor=7; rNsEucl=4; rSSD=2; rNSW=3; rank_mean=5.000; match_rank=5
a 0 0 0 0 0 0 0 15 1 0 39 23 16 0 0 0
c 0 0 0 0 0 0 0 17 0 1 0 0 4 0 0 0
g 0 0 0 0 0 0 0 3 39 39 1 0 16 0 0 0
t 0 0 0 0 0 0 0 5 0 0 0 17 4 0 0 0
|
| MA0080.2_shift7 (SPI1) |
 |
0.842 |
0.453 |
5.422 |
0.442 |
0.895 |
1.081 |
0.923 |
7 |
5 |
5 |
4 |
7 |
5 |
5 |
5.429 |
6 |
; oligos_7nt_mkv4_m3 versus MA0080.2 (SPI1); m=6/8; ncol2=7; w=7; offset=6; strand=D; shift=7; score= 5.4286; -------AGGAAGT--
; cor=0.842; Ncor=0.453; logoDP=5.422; NIcor=0.442; NsEucl=0.895; SSD=1.081; NSW=0.923; rcor=7; rNcor=5; rlogoDP=5; rNIcor=4; rNsEucl=7; rSSD=5; rNSW=5; rank_mean=5.429; match_rank=6
a 0 0 0 0 0 0 0 33 4 4 42 41 5 3 0 0
c 0 0 0 0 0 0 0 3 0 0 0 0 3 5 0 0
g 0 0 0 0 0 0 0 6 37 38 0 1 33 2 0 0
t 0 0 0 0 0 0 0 0 1 0 0 0 1 32 0 0
|
| MA0062.2_shift6 (GABPA) |
 |
0.845 |
0.422 |
7.243 |
0.402 |
0.897 |
1.360 |
0.915 |
6 |
7 |
2 |
5 |
6 |
7 |
7 |
5.714 |
7 |
; oligos_7nt_mkv4_m3 versus MA0062.2 (GABPA); m=7/8; ncol2=11; w=8; offset=5; strand=D; shift=6; score= 5.7143; ------CCGGAAGygv
; cor=0.845; Ncor=0.422; logoDP=7.243; NIcor=0.402; NsEucl=0.897; SSD=1.360; NSW=0.915; rcor=6; rNcor=7; rlogoDP=2; rNIcor=5; rNsEucl=6; rSSD=7; rNSW=7; rank_mean=5.714; match_rank=7
a 0 0 0 0 0 0 32 70 0 0 991 989 94 56 154 264
c 0 0 0 0 0 0 768 914 0 0 1 2 32 261 137 262
g 0 0 0 0 0 0 188 4 990 991 1 0 866 37 603 415
t 0 0 0 0 0 0 1 1 1 0 0 2 0 637 96 49
|
| MA0136.1_rc_shift6 (ELF5_rc) |
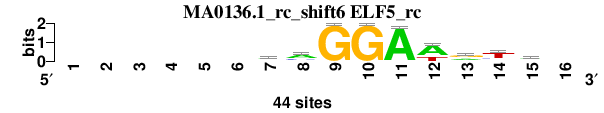 |
0.758 |
0.433 |
0.210 |
-0.127 |
0.888 |
1.608 |
0.899 |
8 |
6 |
7 |
8 |
8 |
8 |
8 |
7.571 |
8 |
; oligos_7nt_mkv4_m3 versus MA0136.1_rc (ELF5_rc); m=8/8; ncol2=9; w=8; offset=5; strand=R; shift=6; score= 7.5714; ------mmGGAwrtw-
; cor=0.758; Ncor=0.433; logoDP=0.210; NIcor=-0.127; NsEucl=0.888; SSD=1.608; NSW=0.899; rcor=8; rNcor=6; rlogoDP=7; rNIcor=8; rNsEucl=8; rSSD=8; rNSW=8; rank_mean=7.571; match_rank=8
a 0 0 0 0 0 0 21 26 0 0 43 29 17 5 20 0
c 0 0 0 0 0 0 11 11 0 0 0 1 5 7 3 0
g 0 0 0 0 0 0 8 3 44 44 1 0 20 3 9 0
t 0 0 0 0 0 0 4 4 0 0 0 14 2 29 12 0
|








